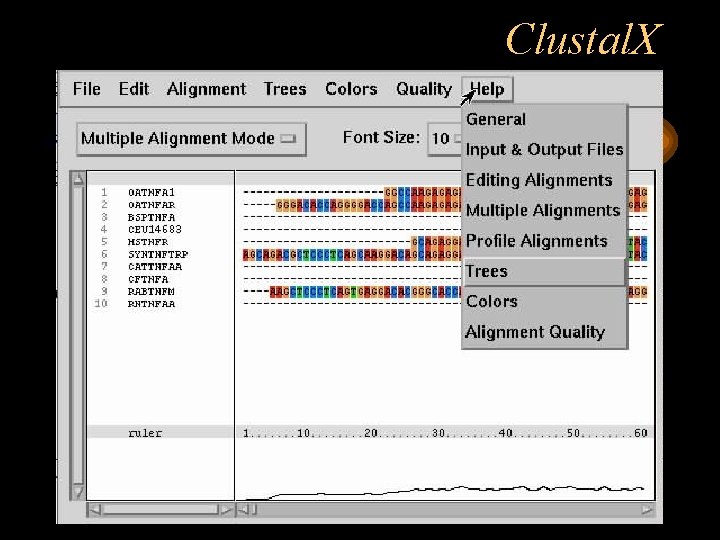
Summary: The Clustal W and Clustal X multiple sequence alignment programs have been completely rewritten in C. This will facilitate the further development of the alignment algorithms in the future and has allowed proper porting of the programs to the latest versions of Linux, Macintosh and Windows operating systems. Clustal X is a windows interface for the ClustalW multiple sequence alignment program. It provides an integrated environment for performing multiple sequence and profile alignments and analysing the results. The sequence alignment is displayed in a window on the screen. A versatile coloring scheme has been incorporated allowing you to highlight. Clustal X help to the Bioinformatics candidate to predicts the Multiple Sequence Alignment and Phylogenetic Analysis for given a nuber of Gene Sequences of varrious organism,and find the evolutionary relationship. Clustal X Colour Scheme This is an emulation of the default colourscheme used for alignments in Clustal X, a graphical interface for the ClustalW multiple sequence alignment program. Each residue in the alignment is assigned a colour if the amino acid profile of the alignment at that position meets some minimum criteria specific for the residue.
This video describes how to perform a multiple sequence alignment using the ClustalX software. The video also discusses the appropriate types of sequence dat.
Introduction
Clustal X is essentially the same program as Clustal W. It is a program designed to compute the alignment of multiple sequences of genetic or protein data. This program uses a GUI to work with the program as opposed to Clustal W, which just uses the command line as an interface.
Using Clustal X on RCC Resources
Clustal X, much like its sister program Clustal W, does not require a module to be loaded in order to run. Clustal X is the GUI version of Clustal W so it is primarily designed to be an interactive program. In order to start the GUI for Clustal X, simply run clustalx from the command line and the GUI will start.
Clustal X (Thompson et al. 1997) is a version of Clustal W with a graphical user interface. The current version is Clustal X2 (Larkin et al. 2007). The program is designed to (1) perform multiple alignments, (2) view the results of the alignment process, and (3) if necessary, improve the alignment. Improving the alignment is facilitated by options that are not available in Clustal W (e.g., selecting a section of the alignment to be realigned with different gap penalties, while keeping the rest of the alignment fixed).

The method Clustal uses to construct the alignment is called pairwise progressive sequence alignment. This heuristic method first does a pairwise sequences alignment for all the sequence pairs that can be constructed from the sequence set. A dendrogram (guide tree) of the sequences is then done according to the pairwise similarity of the sequences. Finally a multiple sequence alignment is constructed by aligning sequences in the order, defined by the guide tree.
Input Format:
To run Clustal X2 in the terminal, type:
Clustalx2 Download
clustalxClustal X2 is available for several computing platforms.

Clustalx2

From the Clustal X window you can use the pull-down menus to import sequences, align sequences, save sequences or alignments, and perform other program tasks.
Download Clustal W2 (clustalx-windows-xp-2.0.zip) and open:
Download Clustal W2 (clustalx-mac-universal-2.0.tgz) and follow the “Install on Mac OS X” instructions in the file: